Indexing & Abstracting
Full Text
Research ArticleDOI Number : 10.36811/ijdsc.2019.110001Article Views : 18Article Downloads : 23
Identification of Trichophyton mentagrophytes strains isolated from patients with dermatophytosis
Imen Dhib1*, Yaacoub A1, Ben Said M1, Fathallah A1 and Zemni R2
1Parasitology-Mycology Laboratory, Farhat Hatched Hospital, Sousse, Tunisia
2Medicine Faculty, Immunology and Genetic Laboratory, Sousse, Tunisia
*Corresponding Author: Imen Dhib, Faculty of Medicine, Parasitology-Mycology laboratory, Mohamed El Karoui Street, 4002 Sousse, Tunisia, Tel: +216 73 222 600; Fax: +216 73 224 899; Email: dhib.imen@yahoo.fr
Article Information
Aritcle Type: Research Article
Citation: Imen Dhib, Yaacoub A, Ben Said M, et al. 2019. Identification of Trichophyton mentagrophytes strains isolated from patients with dermatophytosis. Int J Dermatol Skin Care. 1: 01-07.
Copyright: This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Copyright © 2019; Imen Dhib
Publication history:
Received date: 25 May, 2019Accepted date: 24 June, 2019
Published date: 26 June, 2019
Abstract
According to epidemiological, clinical and mycological criteria, it has long been admitted that the Trichophyton mentagrophytes species includes two varieties: a zoophilic variety (var. mentagrophytes) and an anthropophilic variety (var. interdigital) that involve the upper and the lower part of the body respectively. The further application of molecular techniques to the characterization of dermatophyte strains showed that this classification is unreliable. The aim of our study was to assess the usefulness of PCR-RFLP (restriction fragment length polymorphism) and sequencing in the characterization of T. mentagrophytes strains taken from Tunisian patients. The study was carried out in 2008 in the laboratory of Parasitology-Mycology of Farhat Hatched hospital, Sousse, Tunisia. A total of 133 strains were isolated from 133 patients addressed to the laboratory for dermatological lesions very evocative of dermatomycosis. Eighty strains were isolated from lesions located on the lower part of the body (onychomycosis, tinea pedis) and 53 strains from the upper part of the body (tinea capitis, tinea corporis). All strains were submitted to mycological examination (direct microscopic examination, and culture on Sabered medium) and further investigated by using RFLP analysis of the PCR amplified ITS1-5.8 s and ITS2 region of the ribosomal DNA and the MvaI restriction enzyme. In addition, 20 strains were further submitted to a sequencing of the ITS1-5.8 s and ITS2 region. On the basis of mycological criteria all strains were diagnosed as T. mentagrophytes. All strains produced the same RFLP pattern and were identified as T. mentagrophytes interdigital regardless of the location of lesions. Out of the 20 sequenced strains, five were found anthropophilic and 15 were zoophilic. In conclusion, all strains provisionally diagnosed as T. mentagrophytes on the basis of mycological criteria were shown to belong to T. interdigital by using PCR-RFLP and sequencing irrespective of the site of lesions. The predominance of zoophilic strains needs further investigation.
Keywords: Dermatophytes; Trichophyton mentagrophytes; PCR-RFLP; Sequencing; Phenotypic features
Introduction
Dermatophytes are the main causal agents of superficial mycoses in humans and animals. They are usually identified on the basis of macroscopic appearance, together with microscopic examination of cultures. According to epidemiological, clinical and mycological criteria, it has long been admitted that the T. mentagrophytes species includes two varieties: a zoophilic variety (var. mentagrophytes) and an anthropophilic variety (var. interdigital) that involve the upper and the lower part of the body respectively [1-3]. The further development of molecular tools and techniques for the characterization of strains of the T. mentagrophytes compel [4] and the comparison between phenotypic and genotypic findings [5] showed that this classification was not appropriate [6,7]. The aim of our study was to assess the usefulness of PCR-RFLP (restriction fragment length polymorphism) and sequencing in the identification of T. mentagrophytes strains taken from different body lesions in patients originating from Tunisia.
Material and Methods
Fungal strains
The study was carried out in 2008 in the laboratory of Parasitology-Mycology of Farhat Hatched hospital, Sousse, Tunisia. It included 133 strains of the T. mentagrophytes complex: 129 were isolated from 129 patients originating from Sousse, addressed to the laboratory for dermatological lesions very evocative of dermatomycosis; 2 strains from 2 patients originating from Tunis (Northern Tunisia) and 2 from Sfax (Southern Tunisia). Two reference strains from CBS center (Central Bureau voor Schimmel cultures CBS Utrecht, Netherlands) were included in the study: T. interdigital CBS 165.66 and T. mentagrophytes CBS 106.67. Fungal strains were divided into two groups: 80 strains were isolated from lesions located on the lower part of the body (toes onychomycosis, tinea pedis), designed Ginf and 53 strains from the upper part of the body (tinea capitis, tinea corporis), designed Gsup.
Morphological identification
The identification of T. mentagrophytes strains was made on the basis of macroscopic and microscopic growth criteria. All isolates were cultured in tubes on Sabered agar with chloramphenicol (0.5g/l) and cycloheximide (0.5 g/l) at 27° for 3 weeks. The cultures were examined for macroscopic characters including texture colony and pigmentation. Microscopic examination was performed according to the standard procedure using adhesive tape and lactophenol blue stain. Strains were identified as members of the T. mentagrophytes complex according to the following criteria: powdery to cottony texture, color of colonies and reverse pigmentation, production of round to broadly clavate microconidia, formation of cigar shaped macroconidia and spiral hyphae.
Polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP) of ITS regions
Extraction of genomic DNA was performed using the rapid mini preparation method previously described by Liu [8] with minor modifications. In brief, a small lump of mycelia was added to 500 μl of lysis buffer (400 mM Tris-HCL pH 8; 60 mM EDTA pH 8; 150 mM Nacl; 1% SDS) and 150 μl of potassium acetate 5M. The tube was vortexed and centrifuged at 6000t/mn x3 mn. Isopropyl alcohol was added to the supernatant (w/w) and the solution was centrifuged at 6000t/mn x3 mn. The resultant DNA pellet was washed in 70% ethanol and dissolved in50 μl Tris-EDTA buffer. PCR was performed using the universal primers Mas 266 (5’ GCA TTC CCA AAC TCG ACTC 3’) and V9D (5’ TTA CGT CCC TGC CCT TTG TA 3’) that amplify a DNA fragment of approximately 1Kb. For RFLP, the amplicon was digested with the restriction enzyme MvaI (Promega, Madison WI, USA) for 2 hours at 60°. The resulting restriction fragments were separated electrophoretic ally on 3% agarose gel for 60V at 90 min and visualized under UV light after ethidium bromide staining. Gel photography was obtained using camera (Biometric, Germany).
Sequencing
Out of the 133 strains, 20 were submitted to sequencing of the ITS1-5.8S and ITS2 regions ITS (Internal Transcribed Spacer) of rDNA. They include 7 strains from toes onychomycosis, 2 from tinea pedis, 4 from tinea capitis and 7 from tinea corporis. Sixteen isolates were from patients living in Sousse, 2 in Sfax and 2 in Tunis. Animal contact could be documented for 13 of them: 2 with cats, 1 with chicken, 4 with dogs, 3 with rabbits and 3 with sheep. PCR products were purified using the Big-Dye® Terminator v1.1 cycle sequencing kit (Applied Biosystems) [9].
Statistical analysis
In order to compare the phenotypic and genotypic features of T. mentagrophytes strains, all the collected data were processed in the SPSS database computer program version 11.5 (SPSS Inc, Chicago, USA). The results were analyzed using the chi-squared test. A P value of <0.05% was considered to be significant.
Results
Mycological identification of strains
Detailed morphological characteristics of the 133 strains are shown in Table 1. All clinical isolates from both Ginf and Gsup groups were provisionally identified as T. mentagrophytes on the basis of macroscopic and mycological features. Colonies of most strains were powdery with no difference between both groups (88.1% for Gsup vs 87.5% for Ginf). Colonies yielded by strains of both groups were mostly white (84.9% for Gsup vs 85% for Ginf). The reverse of most strains was colorless or had a yellow pigment. A red to brown reverse pigment was present in 20.7% and 31.1% of strains of groups Gsup and Ginf respectively. Microconidia were abundant to very abundant in strains of both groups. They were spherical in 77.3% and 66.3% of strains of Gsup and Ginf respectively. Macroconidia were present in nearly 50% of strains (45.4% for Gsup vs 42.5% for Ginf). They were abundant in 35.8% of Gsup strains and 30% of Ginf strains. Thus, the phenotypic features of strains isolated from Gsup and Ginf were very similar.
RFLP and sequencing
All the 133 strains gave the expected 1 kb band and produced the same RFLP pattern after digestion with MvaI composed of five bands Figure 1. This pattern is identical to that of the T. interdigital CBS165.66 reference strain, regardless of the phenotypic features and the location of lesions (Gsup or Ginf). The sequences of all 20 strains showed an identity with the ITS regions of the Interdigital strain CBS165.66 registered in NCBI database. On the other hand, the sequence alignment of the ITS regions of 5 T. mentagrophytes strains (2 from Tunis, 2 from Sfax and 1 from Sousse) showed 100% homology with the anthropophilic T. interdigital AF506033 strain.
The sequence alignment of 15 T. mentagrophytes strains from Sousse showed 100% homology with the zoophilic type III* T. interdigital strain FM986758. The sequences of all 20 strains are registered in GenBank with accession numbers from KU921371to KU921390.
|
Table 1: Phenotypic features of 133 T. mentagrophytes strains: 53 from Gsup lesions and 80 from Ginf lesions. |
||||
|
Colony |
Lesion site morphology |
Gsup N(%) |
Ginf N(%) |
P |
|
Texture of colonies |
Powdery |
47 (88.7) |
70 (87.5) |
|
|
Cottony |
6 (11.3) |
10 (12.5) |
|
|
|
|
|
|
|
|
|
|
|
|
||
|
Color of colonies |
White |
45 (84.9) |
68 (85) |
|
|
Cream |
8 (15.1) |
12 (15) |
|
|
|
Reverse pigment |
Red to brown |
11 (20.7) |
25 (31.1) |
|
|
Colorless to yellow |
42 (79.3) |
55 (68.7) |
|
|
|
Microconidia shape |
Spherical |
41 (77.3) |
53 (66.3) |
|
|
Pyriform+ spherical |
12 (22.7) |
27 (33.7) |
<<<3.84 |
|
|
Abundance of microconidia |
+/ ++ |
15 (28.3) |
27 (33.7) |
(NS) |
|
+++ / 4+ |
38 (71.7) |
53 (66.3) |
|
|
|
Abundance of macroconidia |
Absent |
24(45.4) |
34 (42.5) |
|
|
+/- to + |
10 (18.8) |
22 (27.5) |
|
|
|
++/+++ |
19 (35.8) |
24 (30) |
|
|
|
Spiral hyphae |
Absent |
3 (5.6) |
6 (7.5) |
|
|
Spiral hyphae |
50 (94.4) |
74 (92.5) |
|
|
|
Abundance of spiral hyphae |
+/++ |
40 (80) |
66 (89) |
|
|
+++/++++ |
10 (20) |
8 (11) |
|
|
|
NS: no significant value. +/- to +: rare; +/ ++ or++/+++: abundant; +++ / 4+: very abundant. |
||||
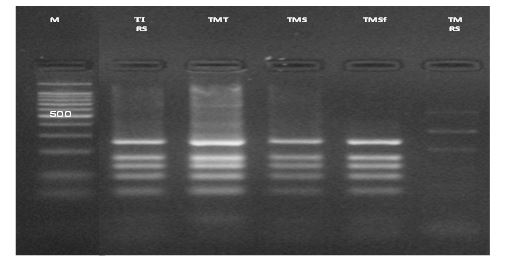
Figure 1: Restriction fragment length polymorphism patterns of internal transcribed spacer- polymerase chain reaction products digested with the restriction enzyme MvaI of T. mentagrophytes. M: molecular size marker 100 pb; TI RS: reference strain of T. interdigitale; TMT: T. mentagrophytes strains from Tunis; TMS: T. mentagrophytes strains from Sousse; TMSf: T. mentagrophytes strains from Sfax; TM RS: reference strain of T. mentagrophytes.
Discussion and Conclusion
In this study, 133 strains of dermatophytes isolated from Tunisian patients with variable dermatophytid lesions, provisionally identified as T. mentagrophytes on the basis of morphological criteria, were further characterized by PCR-RFLP and sequencing of ITS regions which are reported to be the more suitable techniques for the characterization of dermatophyte strains[4,5,10,11]. All typed strains were identified as T. interdigital irrespective of the lesion site. Our findings are in agreement with most recent reports and with the new classification criteria of dermatophytes causing pathology in humans [3,6,12]. The ancient distinction between the strains involving the upper part of the body regarded as zoophilic and named T. mentagrophytes var. mentagrophytes and those involving the lower part of the body considered as anthropophilic and named T. mentagrophytes var. interdigital is no more valid [1,13,14]. So that nearly all human strains of the T. mentagrophytes complex are now considered to belong to T. interdigital species [3,6,15,16]. In our study, mycological characteristics of strains (texture and color of colonies, reverse pigment, abundance of conidia and of spiral hyphae) obtained from lesions located in the upper part of the body (Gsup) and those obtained from lesions located in the lower part of the body (Ginf) were very similar and no statistical difference could be demonstrated between both groups. Our results are in agreement with those of Takashi et al [5] and Nenoff et al [3] who found no relation between morphology of strains and the location of the lesions (lower vs upper part of the body). Our findings are however in contrast with some other previous studies where the T. mentagrophytes var. interdigital strains were reported to be cottony as compared to those of the T. mentagrophytes var. mentagrophytes characterized by a much more granular texture [1-17]. These conflicting results argue for the unreliability of morphological criteria and for the need of molecular techniques for the correct characterization of strains and species.
In our study, no variability between strains was demonstrated as all 133 T. mentagrophytes strains showed the same profile in RFLP which was identical to the T. interdigital CBS165.66 strain. Gupta et al [18] described two different profiles Tm1 and Tm2 by using RFLP of the amplified 18rDNA and ITS regions. Takashi et al [5], divided T. mentagrophytes in 12 types (P1 to P12) by using Southern blot PCR-RFLP of NTS region and established a correlation between types and macroscopic features. At the same time, Ninet et al [19], by sequencing ITS regions of rDNA, identified three types (I, II, III) in T. mentagrophytes strains but without difference in phenotypic traits between the three types. Later, Hedemann et al [7], reported that T. interdigital strains could be divided into 5 different types (I, II, III, III* and IV) on the basis of the sequences of ITS regions; types I and II being anthropophilic and types III, III* and IV being zoophilic. Kim et al [12] reported that RAPD profiles of T. mentagrophytes strains were different, according to the colonies’ texture of studied isolates and showed that colonies of strains of animal origin had a characteristic granular texture in contrast to the anthropophilic strains which were powdery or cottony. Similar findings were reported by Arabizes et al [20] who showed that zoophilic strains isolated from rabbits were phenotypically monomorphic with granular colonies, red brown reverse, a higher number of microconidia and less macroconidia than anthropophilic strains. In contrast, Hedemann et al [7] showed that colonies of T. interdigital strains were either powdery or cottony and this, in the same proportion, whatever they were anthropophilic or zoophilic; and no relation between macroscopic characteristics and origin of strains was demonstrated. Our results are similar to those of Hedemann et al [7] as the majority of our strains had a powdery texture irrespective of their origin, which could only be determined by sequencing. Kac et al [2], Kim et al [12] and Takashi et al [5] found no differences between strains ‘profiles in PCR RFLP and RAPD when lesion location was considered. In contrast, ninety et al [19] showed that toes’ onychomycosis and tinea pedis are caused by type I and II strains while lesions of the upper part of the body are caused by strains of type III. Similar findings were reported by Hedemann et al [7] who showed that lesions of the upper part of the body are caused by the zoophilic type III and III* strains while the anthropophilic strains of type I and II mainly causes toes ’onychomycosis and tinea pedis. This study showed for the first time that tinea corporis and tinea pedis/ onychomycosis are caused by ecologically different strains (zoophilic and anthropophilic origin). In our study, the sequencing of 20 strains showed that 5 were anthropophilic and 15were zoophilic and that there was no relationship between the origin of strains on one hand and the lesion site and the macroscopic features on the other hand. In conclusion, our study confirms that mycological criteria are not reliable for the characterization of strains of the T. mentagrophytes complex and that PCR RFLP and sequencing are much more appropriate for this purpose. Actually, all of our strains identified as T. mentagrophytes on the basis of morphological criteria, were shown to belong to the T. interdigital species whatever the site of lesion.
Acknowledgment
This work was supported by the 04/UR/08-05.Research Unit, from the Ministry of Health, Tunisia.
References
1. Badillet G. 1994. Dermatophytosis and dermatophytes. Encycl. Méd. Chir. (Paris-France), maladies infectieuses.
2. Kac G, Bougnoux ME, Feuilhade DCM, et al. 1999. Genetic diversity among Trichophyton mentagrophytes isolates using random amplified polymorphic DNA method. BR J Dermatol. 140: 839-844. Ref.: https://bit.ly/2WXNCBY
3. Nenoff P, Herrmann J, Gräser Y. 2007. Trichophyton mentagrophytes sive interdigitale? A dermatophyte in course of time. J Dtsch Dermatol Ges. 5: 198-202. Ref.: https://bit.ly/2L7ZPSl
4. Makimura K, Takashi M, Atsuhiko H, et al. 1998. Phylogenetic classification of Trichophyton mentagrophytes complex strains based on DNA sequences of nuclear ribosomal internal transcribed spacer 1 region. J Clin Microbiol. 36: 29-33. Ref.: https://bit.ly/2L7wFmn
5. Takashi M, Ishizaki H, Barton RC, et al. 2003. Restriction fragment length polymorphism analysis of ribosomal DNA intergenic regions is useful for differentiating strains of Trichophyton mentagrophytes. J Clin Microbiol. 41: 4583-4588. Ref.: https://bit.ly/2KwTh05
6. Gräser Y, Scott J, Summerbel R. 2008. The new species concept in dermatophytes- a polyphasic approach. Mycopathologia. 166: 239-256. Ref.: https://bit.ly/2Kx4bDk
7. Heidmann S, Monod M, Gräser Y. 2010. Signature polymorphisms in the internal transcribed spacer region relevant for the differentiation of zoophilic and anthropophilic strains of Trichophyton interdigitale and other species of T mentagrophytes. Br J Dermatol. 162: 282-295. Ref.: https://bit.ly/2xah1hU
8. Liu D, Coloe S, Baird R, et al. 2000. Rapid Mini-preparation of fungal DNA for PCR. J. Clin. Microbiol. 38: 471. Ref.: https://bit.ly/2Y49a1d
9. Tatusova TA, Madden TL. 1999. Blast 2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 174: 247-250. Ref.: https://bit.ly/2L7PrKp
10. Monod M, Bontems O, Zaugg C, Léchenne B, Fratti M, Panizzon R. Fast and reliable PCR/sequencing/RFLP assay for identification of fungi in onychomycoses. J Med Microbiol. 2006; 55:1211-1216
11. White T C, Oliver B G, Gräser Y, et al. 2008. Generating and testing molecular hypotheses in the dermatophytes. Eukaryotic Cell. 7: 1238-1245. Ref.: https://bit.ly/2xb3mr6
12. Kim JA, Takahashi Y, Tanaka R, et al. 2001. Identification and subtyping of Trichophyton mentagrophytes random amplified polymorphic DNA. Mycoses.44: 175-165. Ref.: https://bit.ly/2Y5zuIq
13. Weitzman I, Arvind A. 1996. Dermatophytes: Gross and Microscopic. In: Editor L. Mervyn, MD. Elgart. Dermatologic Clinics, cutaneous mycology. W.B Saunders company. 14: 9-12.
14. Gayle D, Masri-Fridling. 1996. Dermatophytosis of the feet. In: Editor L. Mervyn, MD. Elgart. Dermatologic Clinics, cutaneous mycology. W.B Saunders company. 14: 33-36.
15. Mochizuki T, Watanabe S, Uehara M. 1996. Genetic homogeneity of Trichophyton mentagrophytes var. interdigital isolated from geographically distant regions. J Med Vet Mycol.34: 139-143. Ref.: https://bit.ly/2KubATL
16. De Baere T, Summerbell R, Theelen B, et al. 2010. Evaluation of internal spacer 2-RFLP analysis for the identification of dermatophytes. J Med Microbiol.59: 48-54. Ref.: https://bit.ly/2RqOFt4
17. Koenig H. 1995. Dermatophytes in “Medical mycology instructions”. Ellipse-Edition. 97-134.
18. Gupta AK, Kohli Y, Summerbell RC. 2001. Variation in restriction fragment Length polymorphism among serial isolates from patients with Trichophyton rubrum infection. J Clin Microbiol.39: 260-266. Ref.: https://bit.ly/2N3ibGU
19. Ninet B, Jan I, Bontems O et al. 2003. Identification of dermatophyte species by 28S ribosomal DNA sequencing with a commercial Kit. J Clin Microbiol. 41: 826-830. Ref. : https://bit.ly/2Y6TWZi
20. Arabatzis M, Xylouri E, Frangiadaki I et al. 2006. A Rapid detection of Arthroderma vanbreuseghemii in rabbit skin specimens by PCR-RFLP. Eur Soc Vet Dermatol. 17: 322-326. Ref.: https://bit.ly/2x90GKp
21. Van Rooij P, Detandt M, Nolard N. 2006. Trichophyton mentagrophytes of rabbit origin causing family incidence of kerion: an environmental study. Mycoses. 49: 426-430. Ref.: https://bit.ly/2L7DPXN




















